25. Likelihood Ratio Processes and Bayesian Learning#
25.1. Overview#
This lecture describes the role that likelihood ratio processes play in Bayesian learning.
As in this lecture, we’ll use a simple statistical setting from this lecture.
We’ll focus on how a likelihood ratio process and a prior probability determine a posterior probability.
We’ll derive a convenient recursion for today’s posterior as a function of yesterday’s posterior and today’s multiplicative increment to a likelihood process.
We’ll also present a useful generalization of that formula that represents today’s posterior in terms of an initial prior and today’s realization of the likelihood ratio process.
We’ll study how, at least in our setting, a Bayesian eventually learns the probability distribution that generates the data, an outcome that rests on the asymptotic behavior of likelihood ratio processes studied in this lecture.
We’ll also drill down into the psychology of our Bayesian learner and study dynamics under his subjective beliefs.
This lecture provides technical results that underly outcomes to be studied in this lecture and this lecture and this lecture.
We’ll begin by loading some Python modules.
import matplotlib.pyplot as plt
plt.rcParams["figure.figsize"] = (11, 5) #set default figure size
import numpy as np
from numba import vectorize, jit, prange
from math import gamma
import pandas as pd
import seaborn as sns
colors = sns.color_palette()
@jit
def set_seed():
np.random.seed(142857)
set_seed()
25.2. The Setting#
We begin by reviewing the setting in this lecture, which we adopt here too.
A nonnegative random variable
Before the beginning of time, nature once and for all decides whether she will draw a sequence of IID draws from
We will sometimes let
Nature knows which density it permanently draws from, but we the observers do not.
We do know both
But we want to know.
To do that, we use observations.
We observe a sequence
We want to use these observations to infer whether nature chose
A likelihood ratio process is a useful tool for this task.
To begin, we define the key component of a likelihood ratio process, namely, the time
We assume that
That means that under the
A likelihood ratio process for sequence
where
Sometimes for shorthand we’ll write
Notice that the likelihood process satisfies the recursion or multiplicative decomposition
The likelihood ratio and its logarithm are key tools for making inferences using a classic frequentist approach due to Neyman and Pearson [Neyman and Pearson, 1933].
We’ll again deploy the following Python code from this lecture that
evaluates
# Parameters in the two beta distributions.
F_a, F_b = 1, 1
G_a, G_b = 3, 1.2
@vectorize
def p(x, a, b):
r = gamma(a + b) / (gamma(a) * gamma(b))
return r * x** (a-1) * (1 - x) ** (b-1)
# The two density functions.
f = jit(lambda x: p(x, F_a, F_b))
g = jit(lambda x: p(x, G_a, G_b))
@jit
def simulate(a, b, T=50, N=500):
'''
Generate N sets of T observations of the likelihood ratio,
return as N x T matrix.
'''
l_arr = np.empty((N, T))
for i in range(N):
for j in range(T):
w = np.random.beta(a, b)
l_arr[i, j] = f(w) / g(w)
return l_arr
We’ll also use the following Python code to prepare some informative simulations
l_arr_g = simulate(G_a, G_b, N=50000)
l_seq_g = np.cumprod(l_arr_g, axis=1)
l_arr_f = simulate(F_a, F_b, N=50000)
l_seq_f = np.cumprod(l_arr_f, axis=1)
25.3. Likelihood Ratio Process and Bayes’ Law#
Let
The likelihood ratio process is a principal actor in the formula that governs the evolution
of the posterior probability
Bayes’ law implies that
with
Below we define a Python function that updates belief
@jit
def update(π, l):
"Update π using likelihood l"
# Update belief
π = π * l / (π * l + 1 - π)
return π
Formula (25.1) can be generalized by iterating on it and thereby deriving an
expression for the time
To begin, notice that the updating rule
implies
Therefore
Since
After rearranging the preceding equation, we can express
Formula (25.2) generalizes formula (25.1).
Formula (25.2) can be regarded as a one step revision of prior probability
Formula (25.2) shows the key role that the likelihood ratio process
Formula (25.2) is the foundation for the insight that, because of how the likelihood ratio process behaves
as
To illustrate this insight, below we will plot graphs showing one simulated
path of the likelihood ratio process
First, we tell Python two values of
π1, π2 = 0.2, 0.8
Next we generate paths of the likelihood ratio process
T = l_arr_f.shape[1]
π_seq_f = np.empty((2, T+1))
π_seq_f[:, 0] = π1, π2
for t in range(T):
for i in range(2):
π_seq_f[i, t+1] = update(π_seq_f[i, t], l_arr_f[0, t])
fig, ax1 = plt.subplots()
for i in range(2):
ax1.plot(range(T+1), π_seq_f[i, :], label=fr"$\pi_0$={π_seq_f[i, 0]}")
ax1.set_ylabel("$\pi_t$")
ax1.set_xlabel("t")
ax1.legend()
ax1.set_title("when f governs data")
ax2 = ax1.twinx()
ax2.plot(range(1, T+1), np.log(l_seq_f[0, :]), '--', color='b')
ax2.set_ylabel("$log(L(w^{t}))$")
plt.show()
<>:6: SyntaxWarning: invalid escape sequence '\p'
<>:6: SyntaxWarning: invalid escape sequence '\p'
/tmp/ipykernel_7020/4215127416.py:6: SyntaxWarning: invalid escape sequence '\p'
ax1.set_ylabel("$\pi_t$")
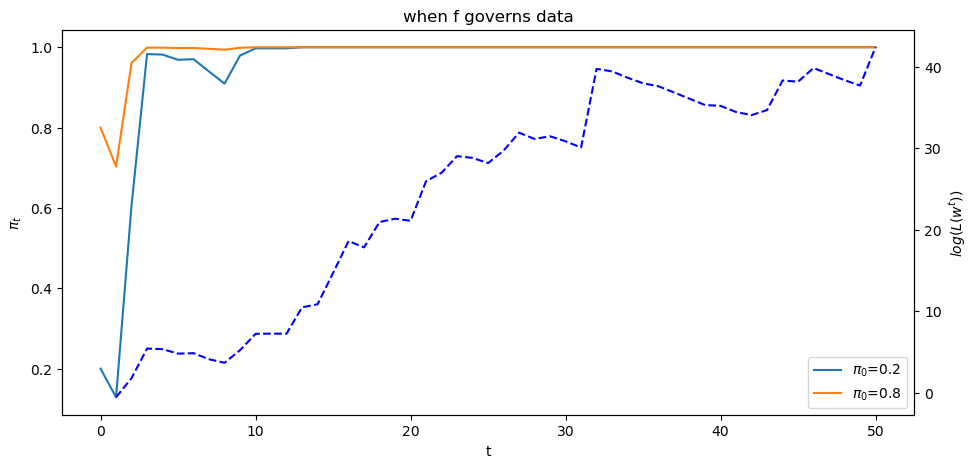
The dotted line in the graph above records the logarithm of the likelihood ratio process
Please note that there are two different scales on the
Now let’s study what happens when the history consists of IID draws from density
T = l_arr_g.shape[1]
π_seq_g = np.empty((2, T+1))
π_seq_g[:, 0] = π1, π2
for t in range(T):
for i in range(2):
π_seq_g[i, t+1] = update(π_seq_g[i, t], l_arr_g[0, t])
fig, ax1 = plt.subplots()
for i in range(2):
ax1.plot(range(T+1), π_seq_g[i, :], label=fr"$\pi_0$={π_seq_g[i, 0]}")
ax1.set_ylabel("$\pi_t$")
ax1.set_xlabel("t")
ax1.legend()
ax1.set_title("when g governs data")
ax2 = ax1.twinx()
ax2.plot(range(1, T+1), np.log(l_seq_g[0, :]), '--', color='b')
ax2.set_ylabel("$log(L(w^{t}))$")
plt.show()
<>:6: SyntaxWarning: invalid escape sequence '\p'
<>:6: SyntaxWarning: invalid escape sequence '\p'
/tmp/ipykernel_7020/1078043107.py:6: SyntaxWarning: invalid escape sequence '\p'
ax1.set_ylabel("$\pi_t$")
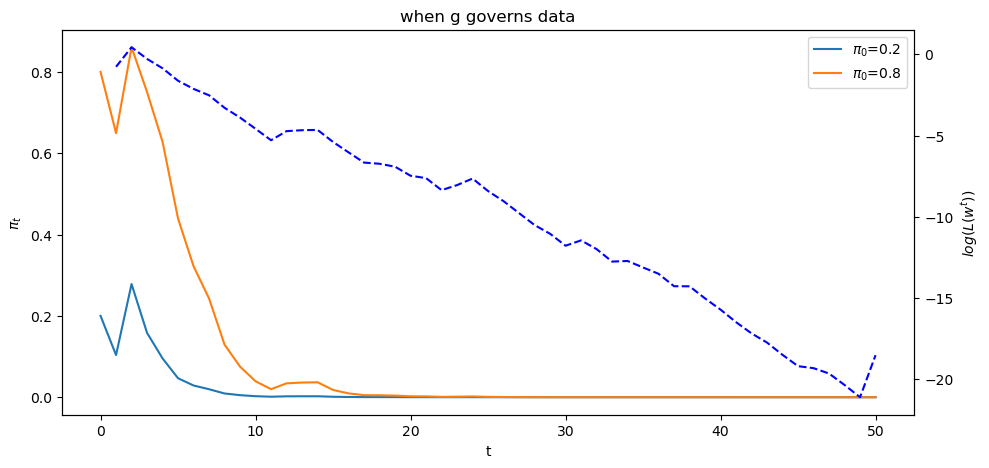
Below we offer Python code that verifies that nature chose permanently to draw from density
π_seq = np.empty((2, T+1))
π_seq[:, 0] = π1, π2
for i in range(2):
πL = π_seq[i, 0] * l_seq_f[0, :]
π_seq[i, 1:] = πL / (πL + 1 - π_seq[i, 0])
np.abs(π_seq - π_seq_f).max() < 1e-10
True
We thus conclude that the likelihood ratio process is a key ingredient of the formula (25.2) for
a Bayesian’s posteior probabilty that nature has drawn history
25.4. Behavior of posterior probability
We’ll end this lecture by briefly studying what our Baysian learner expects to learn under the
subjective beliefs
This will provide us with some perspective on our application of Bayes’s law as a theory of learning.
As we shall see, at each time
But he expects that new information will not lead him to change his beliefs.
And it won’t on average under his subjective beliefs.
We’ll continue with our setting in which a McCall worker knows that successive
draws of his wage are drawn from either
We’ll review and reiterate and rearrange some formulas that we have encountered above and in associated lectures.
The worker’s initial beliefs induce a joint probability distribution
over a potentially infinite sequence of draws
Bayes’ law is simply an application of laws of
probability to compute the conditional distribution of the
After our worker puts a subjective probability
We assume that the worker also knows the laws of probability theory.
A respectable view is that Bayes’ law is less a theory of learning than a statement about the consequences of information inflows for a decision maker who thinks he knows the truth (i.e., a joint probability distribution) from the beginning.
25.4.1. Mechanical details again#
At time
Before drawing a wage at time
Let
After drawing
More generally, after making the
or
and that the density of
Notice that
so that the process
Indeed, it is a bounded martingale because each
In the first line in the above string of equalities, the term in the first set of brackets
is just
Notice that here we are computing
Because
Practically, this means that probability one is attached to sample paths
According to the theorem, it different sample paths can converge to different limiting values.
Thus, let
We can think of nature as drawing an
The limit points of
By staring at law of motion (25.1) or (25.3) , we can figure out some things about the probability distribution of the limit points
Evidently, since the likelihood ratio
and
Thus, for some realizations,
Now let’s remember that
The law of iterated expectations implies
and in particular
Applying the above formula to
where the mathematical expectation
Since the only two values that
and consequently that
Combining this equation with equation (20), we deduce that
the probability that
Thus, under the worker’s subjective distribution,
25.4.2. Some simulations#
Let’s watch the martingale convergence theorem at work in some simulations of our learning model under the worker’s subjective distribution.
Let us simulate
We’ll plot a large sample of paths.
@jit
def martingale_simulate(π0, N=5000, T=200):
π_path = np.empty((N,T+1))
w_path = np.empty((N,T))
π_path[:,0] = π0
for n in range(N):
π = π0
for t in range(T):
# draw w
if np.random.rand() <= π:
w = np.random.beta(F_a, F_b)
else:
w = np.random.beta(G_a, G_b)
π = π*f(w)/g(w)/(π*f(w)/g(w) + 1 - π)
π_path[n,t+1] = π
w_path[n,t] = w
return π_path, w_path
def fraction_0_1(π0, N, T, decimals):
π_path, w_path = martingale_simulate(π0, N=N, T=T)
values, counts = np.unique(np.round(π_path[:,-1], decimals=decimals), return_counts=True)
return values, counts
def create_table(π0s, N=10000, T=500, decimals=2):
outcomes = []
for π0 in π0s:
values, counts = fraction_0_1(π0, N=N, T=T, decimals=decimals)
freq = counts/N
outcomes.append(dict(zip(values, freq)))
table = pd.DataFrame(outcomes).sort_index(axis=1).fillna(0)
table.index = π0s
return table
# simulate
T = 200
π0 = .5
π_path, w_path = martingale_simulate(π0=π0, T=T, N=10000)
fig, ax = plt.subplots()
for i in range(100):
ax.plot(range(T+1), π_path[i, :])
ax.set_xlabel('$t$')
ax.set_ylabel(r'$\pi_t$')
plt.show()
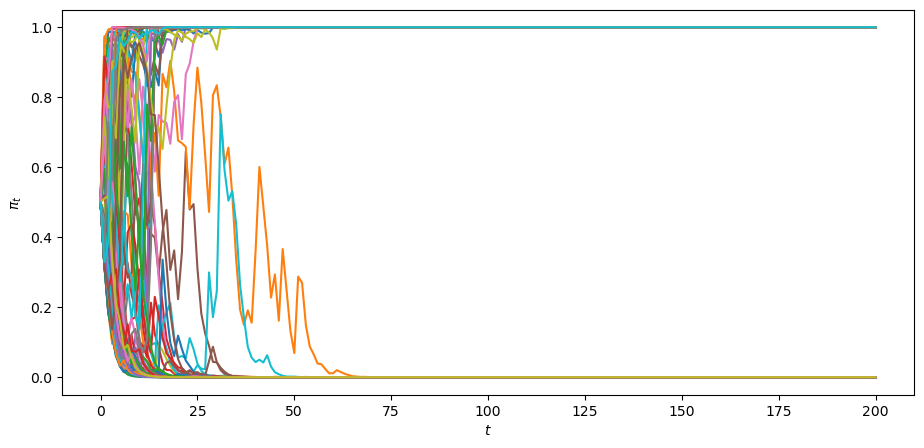
The above graph indicates that
each of paths converges
some of the paths converge to
some of the paths converge to
none of the paths converge to a limit point not equal to
Convergence actually occurs pretty fast, as the following graph of the cross-ensemble distribution of
fig, ax = plt.subplots()
for t in [1, 10, T-1]:
ax.hist(π_path[:,t], bins=20, alpha=0.4, label=f'T={t}')
ax.set_ylabel('count')
ax.set_xlabel(r'$\pi_T$')
ax.legend(loc='lower right')
plt.show()
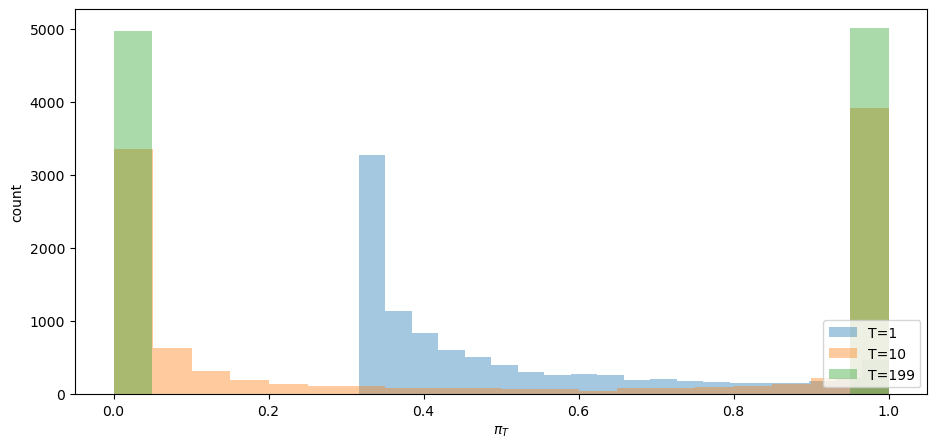
Evidently, by
The fraction of paths that have converged to
The fractions of paths that have converged to
Does the fraction
Yes, it does: it equals the value of
So let’s change
# simulate
T = 200
π0 = .3
π_path3, w_path3 = martingale_simulate(π0=π0, T=T, N=10000)
fig, ax = plt.subplots()
for t in [1, 10, T-1]:
ax.hist(π_path3[:,t], bins=20, alpha=0.4, label=f'T={t}')
ax.set_ylabel('count')
ax.set_xlabel(r'$\pi_T$')
ax.legend(loc='upper right')
plt.show()
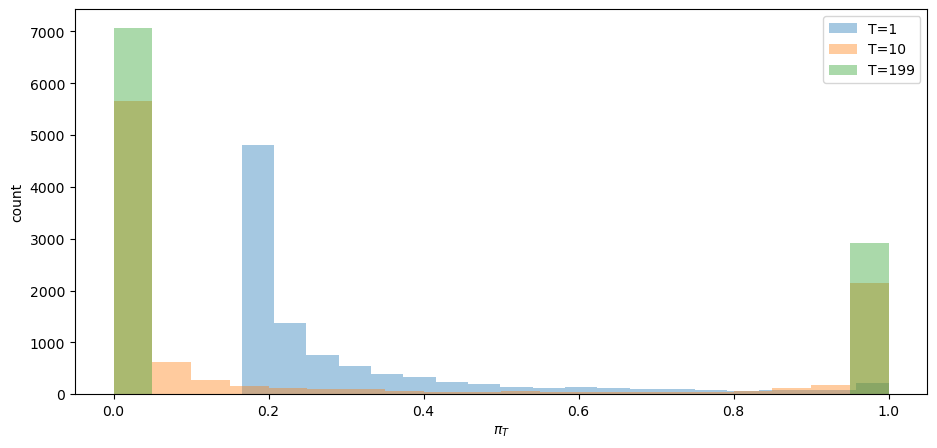
For the preceding ensemble that assumed
Notice that one of the paths involves systematically higher
The luck of the draw early in a simulation push the subjective distribution to draw from
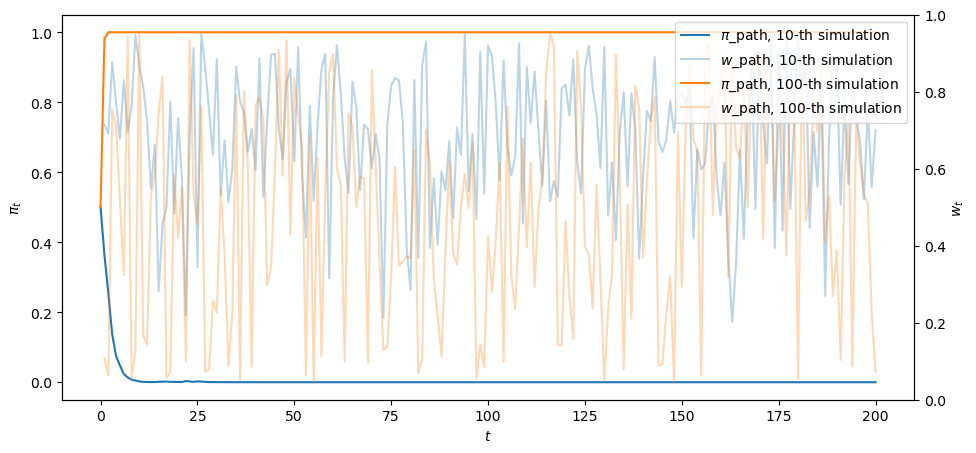
fig, ax = plt.subplots()
for i, j in enumerate([10, 100]):
ax.plot(range(T+1), π_path[j,:], color=colors[i], label=fr'$\pi$_path, {j}-th simulation')
ax.plot(range(1,T+1), w_path[j,:], color=colors[i], label=fr'$w$_path, {j}-th simulation', alpha=0.3)
ax.legend(loc='upper right')
ax.set_xlabel('$t$')
ax.set_ylabel(r'$\pi_t$')
ax2 = ax.twinx()
ax2.set_ylabel("$w_t$")
plt.show()
25.5. Initial Prior is Verified by Paths Drawn from Subjective Conditional Densities#
Now let’s use our Python code to generate a table that checks out our earlier claims about the
probability distribution of the pointwise limits
We’ll use our simulations to generate a histogram of this distribution.
In the following table, the left column in bold face reports an assumed value of
The second column reports the fraction of
The third column reports the fraction of
# create table
table = create_table(list(np.linspace(0,1,11)), N=10000, T=500)
table
| 0.0 | 1.0 | |
|---|---|---|
| 0.0 | 1.0000 | 0.0000 |
| 0.1 | 0.8929 | 0.1071 |
| 0.2 | 0.7994 | 0.2006 |
| 0.3 | 0.7014 | 0.2986 |
| 0.4 | 0.5939 | 0.4061 |
| 0.5 | 0.5038 | 0.4962 |
| 0.6 | 0.3982 | 0.6018 |
| 0.7 | 0.3092 | 0.6908 |
| 0.8 | 0.1963 | 0.8037 |
| 0.9 | 0.0963 | 0.9037 |
| 1.0 | 0.0000 | 1.0000 |
The fraction of simulations for which
25.6. Drilling Down a Little Bit#
To understand how the local dynamics of
Under the subjective distribution this conditional variance is defined as
We can use a Monte Carlo simulation to approximate this conditional variance.
We approximate it for a grid of points
Then we’ll plot it.
@jit
def compute_cond_var(pi, mc_size=int(1e6)):
# create monte carlo draws
mc_draws = np.zeros(mc_size)
for i in prange(mc_size):
if np.random.rand() <= pi:
mc_draws[i] = np.random.beta(F_a, F_b)
else:
mc_draws[i] = np.random.beta(G_a, G_b)
dev = pi*f(mc_draws)/(pi*f(mc_draws) + (1-pi)*g(mc_draws)) - pi
return np.mean(dev**2)
pi_array = np.linspace(0, 1, 40)
cond_var_array = []
for pi in pi_array:
cond_var_array.append(compute_cond_var(pi))
fig, ax = plt.subplots()
ax.plot(pi_array, cond_var_array)
ax.set_xlabel(r'$\pi_{t-1}$')
ax.set_ylabel(r'$\sigma^{2}(\pi_{t}\vert \pi_{t-1})$')
plt.show()
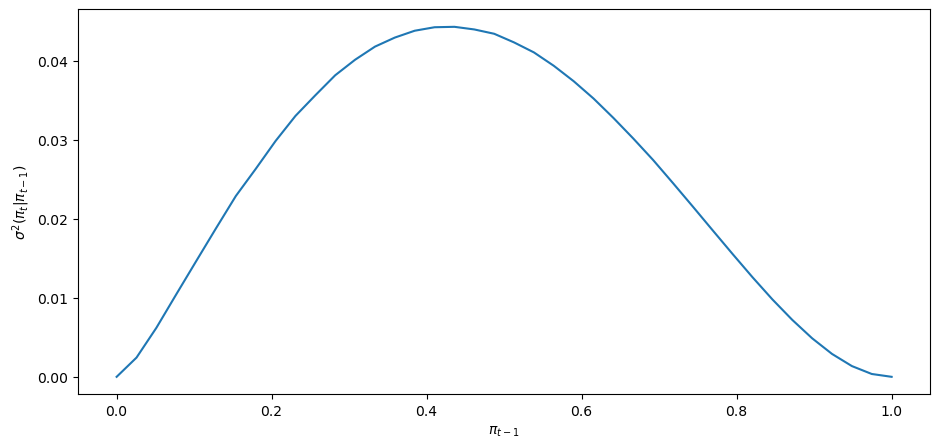
The shape of the the conditional variance as a function of
Notice how the conditional variance approaches
The conditional variance is nearly zero only when the agent is almost sure that
25.7. Sequels#
This lecture has been devoted to building some useful infrastructure that will help us understand inferences that are the foundations of results described in this lecture and this lecture and this lecture.