4. Circulant Matrices#
4.1. Overview#
This lecture describes circulant matrices and some of their properties.
Circulant matrices have a special structure that connects them to useful concepts including
convolution
Fourier transforms
permutation matrices
Because of these connections, circulant matrices are widely used in machine learning, for example, in image processing.
We begin by importing some Python packages
import numpy as np
from numba import jit
import matplotlib.pyplot as plt
np.set_printoptions(precision=3, suppress=True)
4.2. Constructing a Circulant Matrix#
To construct an
After setting entries in the first row, the remaining rows of a circulant matrix are determined as follows:
It is also possible to construct a circulant matrix by creating the transpose of the above matrix, in which case only the first column needs to be specified.
Let’s write some Python code to generate a circulant matrix.
@jit
def construct_cirlulant(row):
N = row.size
C = np.empty((N, N))
for i in range(N):
C[i, i:] = row[:N-i]
C[i, :i] = row[N-i:]
return C
# a simple case when N = 3
construct_cirlulant(np.array([1., 2., 3.]))
array([[1., 2., 3.],
[3., 1., 2.],
[2., 3., 1.]])
4.2.1. Some Properties of Circulant Matrices#
Here are some useful properties:
Suppose that
The transpose of a circulant matrix is a circulant matrix.
Now consider a circulant matrix with first row
and consider a vector
The convolution of vectors
We use
It can be verified that the vector
where
4.3. Connection to Permutation Matrix#
A good way to construct a circulant matrix is to use a permutation matrix.
Before defining a permutation matrix, we’ll define a permutation.
A permutation of a set of the set of non-negative integers
A permutation of a set
A permutation matrix is obtained by permuting the rows of an
Thus, every row and every column contain precisely a single
Every permutation corresponds to a unique permutation matrix.
For example, the
serves as a cyclic shift operator that, when applied to an
Eigenvalues of the cyclic shift permutation matrix
and solving
Eigenvalues
Magnitudes
Thus, singular values of the permutation matrix
It can be verified that permutation matrices are orthogonal matrices:
4.4. Examples with Python#
Let’s write some Python code to illustrate these ideas.
@jit
def construct_P(N):
P = np.zeros((N, N))
for i in range(N-1):
P[i, i+1] = 1
P[-1, 0] = 1
return P
P4 = construct_P(4)
P4
array([[0., 1., 0., 0.],
[0., 0., 1., 0.],
[0., 0., 0., 1.],
[1., 0., 0., 0.]])
# compute the eigenvalues and eigenvectors
𝜆, Q = np.linalg.eig(P4)
for i in range(4):
print(f'𝜆{i} = {𝜆[i]:.1f} \nvec{i} = {Q[i, :]}\n')
𝜆0 = -1.0+0.0j
vec0 = [-0.5+0.j 0. +0.5j 0. -0.5j -0.5+0.j ]
𝜆1 = 0.0+1.0j
vec1 = [ 0.5+0.j -0.5+0.j -0.5-0.j -0.5+0.j]
𝜆2 = 0.0-1.0j
vec2 = [-0.5+0.j 0. -0.5j 0. +0.5j -0.5+0.j ]
𝜆3 = 1.0+0.0j
vec3 = [ 0.5+0.j 0.5-0.j 0.5+0.j -0.5+0.j]
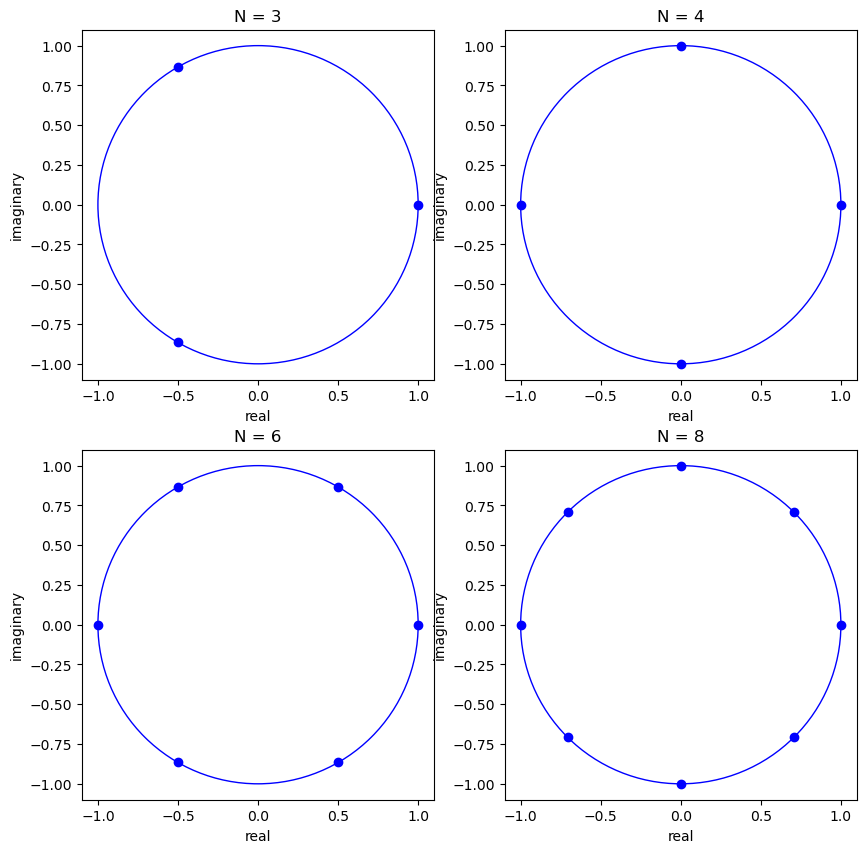
In graphs below, we shall portray eigenvalues of a shift permutation matrix in the complex plane.
These eigenvalues are uniformly distributed along the unit circle.
They are the
In particular, the
where
fig, ax = plt.subplots(2, 2, figsize=(10, 10))
for i, N in enumerate([3, 4, 6, 8]):
row_i = i // 2
col_i = i % 2
P = construct_P(N)
𝜆, Q = np.linalg.eig(P)
circ = plt.Circle((0, 0), radius=1, edgecolor='b', facecolor='None')
ax[row_i, col_i].add_patch(circ)
for j in range(N):
ax[row_i, col_i].scatter(𝜆[j].real, 𝜆[j].imag, c='b')
ax[row_i, col_i].set_title(f'N = {N}')
ax[row_i, col_i].set_xlabel('real')
ax[row_i, col_i].set_ylabel('imaginary')
plt.show()
For a vector of coefficients
Consider an example in which
It can be verified that the matrix
The matrix
To convert it into an orthogonal eigenvector matrix, we can simply normalize it by dividing every entry by
stare at the first column of
The eigenvalues corresponding to each eigenvector are
def construct_F(N):
w = np.e ** (-complex(0, 2*np.pi/N))
F = np.ones((N, N), dtype=complex)
for i in range(1, N):
F[i, 1:] = w ** (i * np.arange(1, N))
return F, w
F8, w = construct_F(8)
w
(0.7071067811865476-0.7071067811865475j)
F8
array([[ 1. +0.j , 1. +0.j , 1. +0.j , 1. +0.j ,
1. +0.j , 1. +0.j , 1. +0.j , 1. +0.j ],
[ 1. +0.j , 0.707-0.707j, 0. -1.j , -0.707-0.707j,
-1. -0.j , -0.707+0.707j, -0. +1.j , 0.707+0.707j],
[ 1. +0.j , 0. -1.j , -1. -0.j , -0. +1.j ,
1. +0.j , 0. -1.j , -1. -0.j , -0. +1.j ],
[ 1. +0.j , -0.707-0.707j, -0. +1.j , 0.707-0.707j,
-1. -0.j , 0.707+0.707j, 0. -1.j , -0.707+0.707j],
[ 1. +0.j , -1. -0.j , 1. +0.j , -1. -0.j ,
1. +0.j , -1. -0.j , 1. +0.j , -1. -0.j ],
[ 1. +0.j , -0.707+0.707j, 0. -1.j , 0.707+0.707j,
-1. -0.j , 0.707-0.707j, -0. +1.j , -0.707-0.707j],
[ 1. +0.j , -0. +1.j , -1. -0.j , 0. -1.j ,
1. +0.j , -0. +1.j , -1. -0.j , 0. -1.j ],
[ 1. +0.j , 0.707+0.707j, -0. +1.j , -0.707+0.707j,
-1. -0.j , -0.707-0.707j, 0. -1.j , 0.707-0.707j]])
# normalize
Q8 = F8 / np.sqrt(8)
# verify the orthogonality (unitarity)
Q8 @ np.conjugate(Q8)
array([[ 1.+0.j, -0.+0.j, -0.+0.j, -0.+0.j, -0.+0.j, 0.+0.j, 0.+0.j,
0.+0.j],
[-0.-0.j, 1.+0.j, -0.+0.j, -0.+0.j, -0.+0.j, -0.+0.j, 0.+0.j,
0.+0.j],
[-0.-0.j, -0.-0.j, 1.+0.j, -0.+0.j, -0.+0.j, -0.+0.j, 0.+0.j,
0.+0.j],
[-0.-0.j, -0.-0.j, -0.-0.j, 1.+0.j, -0.+0.j, -0.+0.j, -0.+0.j,
-0.+0.j],
[-0.-0.j, -0.-0.j, -0.-0.j, -0.-0.j, 1.+0.j, -0.+0.j, -0.+0.j,
-0.+0.j],
[ 0.-0.j, -0.-0.j, -0.-0.j, -0.-0.j, -0.-0.j, 1.+0.j, -0.+0.j,
-0.+0.j],
[ 0.-0.j, 0.-0.j, 0.-0.j, -0.-0.j, -0.-0.j, -0.-0.j, 1.+0.j,
-0.+0.j],
[ 0.-0.j, 0.-0.j, 0.-0.j, -0.-0.j, -0.-0.j, -0.-0.j, -0.-0.j,
1.+0.j]])
Let’s verify that
P8 = construct_P(8)
diff_arr = np.empty(8, dtype=complex)
for j in range(8):
diff = P8 @ Q8[:, j] - w ** j * Q8[:, j]
diff_arr[j] = diff @ diff.T
diff_arr
array([ 0.+0.j, -0.+0.j, -0.+0.j, -0.+0.j, -0.+0.j, -0.+0.j, -0.+0.j,
-0.+0.j])
4.5. Associated Permutation Matrix#
Next, we execute calculations to verify that the circulant matrix
and that every eigenvector of
We illustrate this for
c = np.random.random(8)
c
array([0.279, 0.574, 0.511, 0.431, 0.929, 0.767, 0.175, 0.315])
C8 = construct_cirlulant(c)
Compute
N = 8
C = np.zeros((N, N))
P = np.eye(N)
for i in range(N):
C += c[i] * P
P = P8 @ P
C
array([[0.279, 0.574, 0.511, 0.431, 0.929, 0.767, 0.175, 0.315],
[0.315, 0.279, 0.574, 0.511, 0.431, 0.929, 0.767, 0.175],
[0.175, 0.315, 0.279, 0.574, 0.511, 0.431, 0.929, 0.767],
[0.767, 0.175, 0.315, 0.279, 0.574, 0.511, 0.431, 0.929],
[0.929, 0.767, 0.175, 0.315, 0.279, 0.574, 0.511, 0.431],
[0.431, 0.929, 0.767, 0.175, 0.315, 0.279, 0.574, 0.511],
[0.511, 0.431, 0.929, 0.767, 0.175, 0.315, 0.279, 0.574],
[0.574, 0.511, 0.431, 0.929, 0.767, 0.175, 0.315, 0.279]])
C8
array([[0.279, 0.574, 0.511, 0.431, 0.929, 0.767, 0.175, 0.315],
[0.315, 0.279, 0.574, 0.511, 0.431, 0.929, 0.767, 0.175],
[0.175, 0.315, 0.279, 0.574, 0.511, 0.431, 0.929, 0.767],
[0.767, 0.175, 0.315, 0.279, 0.574, 0.511, 0.431, 0.929],
[0.929, 0.767, 0.175, 0.315, 0.279, 0.574, 0.511, 0.431],
[0.431, 0.929, 0.767, 0.175, 0.315, 0.279, 0.574, 0.511],
[0.511, 0.431, 0.929, 0.767, 0.175, 0.315, 0.279, 0.574],
[0.574, 0.511, 0.431, 0.929, 0.767, 0.175, 0.315, 0.279]])
Now let’s compute the difference between two circulant matrices that we have constructed in two different ways.
np.abs(C - C8).max()
0.0
The
𝜆_C8 = np.zeros(8, dtype=complex)
for j in range(8):
for k in range(8):
𝜆_C8[j] += c[k] * w ** (j * k)
𝜆_C8
array([ 3.982+0.j , -0.868-0.282j, 0.522-0.595j, -0.431+0.391j,
-0.193-0.j , -0.431-0.391j, 0.522+0.595j, -0.868+0.282j])
We can verify this by comparing C8 @ Q8[:, j] with 𝜆_C8[j] * Q8[:, j].
# verify
for j in range(8):
diff = C8 @ Q8[:, j] - 𝜆_C8[j] * Q8[:, j]
print(diff)
[0.+0.j 0.+0.j 0.+0.j 0.+0.j 0.+0.j 0.+0.j 0.+0.j 0.+0.j]
[ 0.+0.j 0.-0.j -0.-0.j -0.-0.j -0.-0.j -0.-0.j -0.+0.j -0.+0.j]
[-0.-0.j -0.-0.j -0.-0.j -0.-0.j -0.-0.j -0.+0.j 0.+0.j 0.-0.j]
[ 0.+0.j -0.-0.j -0.+0.j 0.-0.j -0.-0.j -0.+0.j 0.-0.j -0.-0.j]
[ 0.+0.j -0.-0.j 0.+0.j -0.-0.j 0.-0.j -0.-0.j 0.-0.j -0.-0.j]
[ 0.+0.j -0.-0.j 0.+0.j -0.-0.j 0.-0.j 0.+0.j -0.-0.j 0.-0.j]
[ 0.+0.j -0.-0.j 0.-0.j 0.-0.j 0.-0.j 0.+0.j -0.+0.j -0.-0.j]
[-0.-0.j 0.-0.j 0.-0.j 0.-0.j 0.-0.j 0.-0.j 0.+0.j 0.+0.j]
4.6. Discrete Fourier Transform#
The Discrete Fourier Transform (DFT) allows us to represent a discrete time sequence as a weighted sum of complex sinusoids.
Consider a sequence of
The Discrete Fourier Transform maps
where
def DFT(x):
"The discrete Fourier transform."
N = len(x)
w = np.e ** (-complex(0, 2*np.pi/N))
X = np.zeros(N, dtype=complex)
for k in range(N):
for n in range(N):
X[k] += x[n] * w ** (k * n)
return X
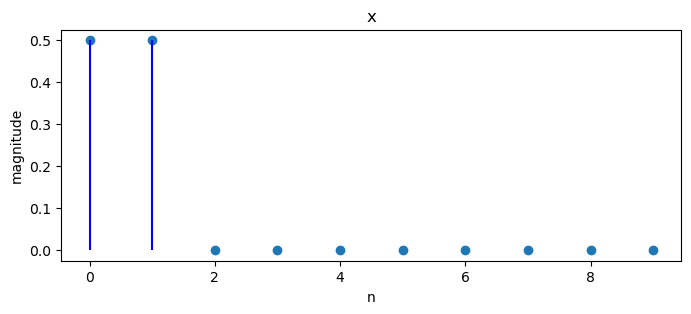
Consider the following example.
x = np.zeros(10)
x[0:2] = 1/2
x
array([0.5, 0.5, 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. ])
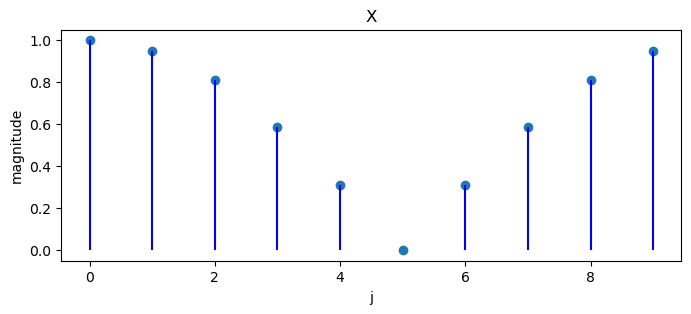
Apply a discrete Fourier transform.
X = DFT(x)
X
array([ 1. +0.j , 0.905-0.294j, 0.655-0.476j, 0.345-0.476j,
0.095-0.294j, -0. +0.j , 0.095+0.294j, 0.345+0.476j,
0.655+0.476j, 0.905+0.294j])
We can plot magnitudes of a sequence of numbers and the associated discrete Fourier transform.
def plot_magnitude(x=None, X=None):
data = []
names = []
xs = []
if (x is not None):
data.append(x)
names.append('x')
xs.append('n')
if (X is not None):
data.append(X)
names.append('X')
xs.append('j')
num = len(data)
for i in range(num):
n = data[i].size
plt.figure(figsize=(8, 3))
plt.scatter(range(n), np.abs(data[i]))
plt.vlines(range(n), 0, np.abs(data[i]), color='b')
plt.xlabel(xs[i])
plt.ylabel('magnitude')
plt.title(names[i])
plt.show()
plot_magnitude(x=x, X=X)
The inverse Fourier transform transforms a Fourier transform
The inverse Fourier transform is defined as
def inverse_transform(X):
N = len(X)
w = np.e ** (complex(0, 2*np.pi/N))
x = np.zeros(N, dtype=complex)
for n in range(N):
for k in range(N):
x[n] += X[k] * w ** (k * n) / N
return x
inverse_transform(X)
array([ 0.5+0.j, 0.5-0.j, -0. -0.j, -0. -0.j, -0. -0.j, -0. -0.j,
-0. +0.j, -0. +0.j, -0. +0.j, -0. +0.j])
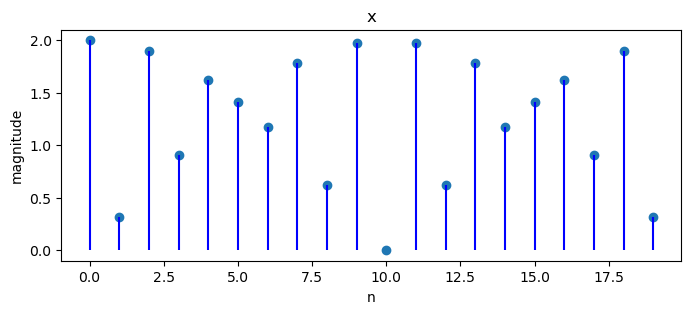
Another example is
Since
To handle this, we shall end up using all
Since
N = 20
x = np.empty(N)
for j in range(N):
x[j] = 2 * np.cos(2 * np.pi * 11 * j / 40)
X = DFT(x)
plot_magnitude(x=x, X=X)
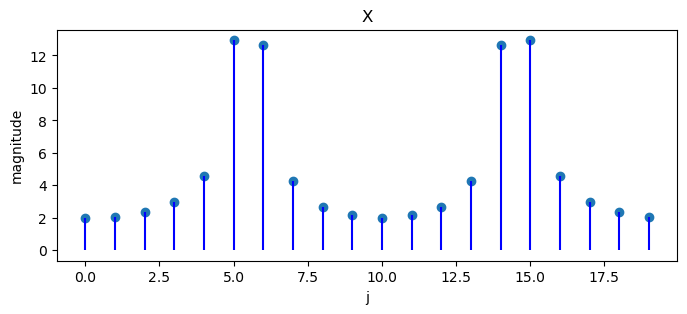
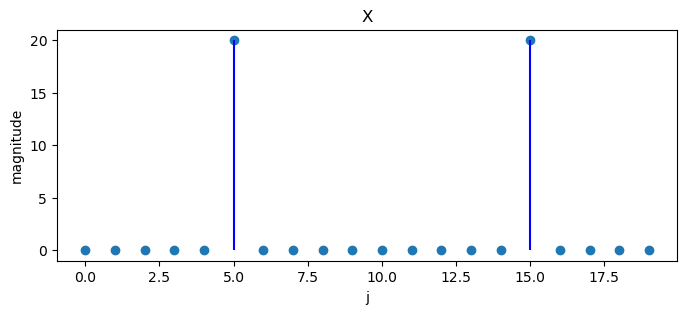
What happens if we change the last example to
Note that
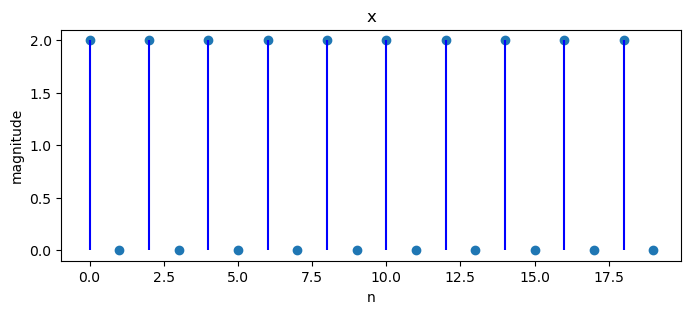
N = 20
x = np.empty(N)
for j in range(N):
x[j] = 2 * np.cos(2 * np.pi * 10 * j / 40)
X = DFT(x)
plot_magnitude(x=x, X=X)
If we represent the discrete Fourier transform as a matrix, we discover that it equals the matrix
We can use the example where
N = 20
x = np.empty(N)
for j in range(N):
x[j] = 2 * np.cos(2 * np.pi * 11 * j / 40)
x
array([ 2. , -0.313, -1.902, 0.908, 1.618, -1.414, -1.176, 1.782,
0.618, -1.975, -0. , 1.975, -0.618, -1.782, 1.176, 1.414,
-1.618, -0.908, 1.902, 0.313])
First use the summation formula to transform
X = DFT(x)
X
array([2. +0.j , 2. +0.558j, 2. +1.218j, 2. +2.174j, 2. +4.087j,
2.+12.785j, 2.-12.466j, 2. -3.751j, 2. -1.801j, 2. -0.778j,
2. -0.j , 2. +0.778j, 2. +1.801j, 2. +3.751j, 2.+12.466j,
2.-12.785j, 2. -4.087j, 2. -2.174j, 2. -1.218j, 2. -0.558j])
Now let’s evaluate the outcome of postmultiplying the eigenvector matrix
F20, _ = construct_F(20)
F20 @ x
array([2. +0.j , 2. +0.558j, 2. +1.218j, 2. +2.174j, 2. +4.087j,
2.+12.785j, 2.-12.466j, 2. -3.751j, 2. -1.801j, 2. -0.778j,
2. -0.j , 2. +0.778j, 2. +1.801j, 2. +3.751j, 2.+12.466j,
2.-12.785j, 2. -4.087j, 2. -2.174j, 2. -1.218j, 2. -0.558j])
Similarly, the inverse DFT can be expressed as a inverse DFT matrix
F20_inv = np.linalg.inv(F20)
F20_inv @ X
array([ 2. -0.j, -0.313-0.j, -1.902+0.j, 0.908-0.j, 1.618-0.j,
-1.414+0.j, -1.176+0.j, 1.782+0.j, 0.618-0.j, -1.975-0.j,
-0. +0.j, 1.975-0.j, -0.618-0.j, -1.782+0.j, 1.176+0.j,
1.414-0.j, -1.618-0.j, -0.908+0.j, 1.902+0.j, 0.313-0.j])