24. Exchangeability and Bayesian Updating#
Contents
24.1. Overview#
This lecture studies learning via Bayes’ Law.
We touch foundations of Bayesian statistical inference invented by Bruno DeFinetti [de Finetti, 1937].
The relevance of DeFinetti’s work for economists is presented forcefully by David Kreps in chapter 11 of [Kreps, 1988].
An example that we study in this lecture is a key component of this lecture that augments the classic job search model of McCall [McCall, 1970] by presenting an unemployed worker with a statistical inference problem.
Here we create graphs that illustrate the role that a likelihood ratio plays in Bayes’ Law.
We’ll use such graphs to provide insights into mechanics driving outcomes in this lecture about learning in an augmented McCall job search model.
Among other things, this lecture discusses connections between the statistical concepts of sequences of random variables that are
independently and identically distributed
exchangeable (also known as conditionally independently and identically distributed)
Understanding these concepts is essential for appreciating how Bayesian updating works.
You can read about exchangeability here.
Because another term for exchangeable is conditionally independent, we want to convey an answer to the question conditional on what?
We also tell why an assumption of independence precludes learning while an assumption of conditional independence makes learning possible.
Below, we’ll often use
Let’s start with some imports:
import matplotlib.pyplot as plt
plt.rcParams["figure.figsize"] = (11, 5) #set default figure size
from numba import jit, vectorize
from math import gamma
import scipy.optimize as op
from scipy.integrate import quad
import numpy as np
24.2. Independently and Identically Distributed#
We begin by looking at the notion of an independently and identically distributed sequence of random variables.
An independently and identically distributed sequence is often abbreviated as IID.
Two notions are involved
independence
identically distributed
A sequence
The sequence
For example, let
Then the joint density of the sequence
so that the joint density is the product of a sequence of identical marginal densities.
24.2.1. IID Means Past Observations Don’t Tell Us Anything About Future Observations#
If a sequence is random variables is IID, past information provides no information about future realizations.
Therefore, there is nothing to learn from the past about the future.
To understand these statements, let the joint distribution of a sequence of random variables
Using the laws of probability, we can always factor such a joint density into a product of conditional densities:
In general,
which states that the conditional density on the left side does not equal the marginal density on the right side.
But in the special IID case,
so that the partial history
So in the IID case, there is nothing to learn about the densities of future random variables from past random variables.
But when the sequence is not IID, there is something to learn about the future from observations of past random variables.
We turn next to an instance of the general case in which the sequence is not IID.
Please watch for what can be learned from the past and when.
24.3. A Setting in Which Past Observations Are Informative#
Let
There are two distinct cumulative distribution functions
Before the start of time, say at time
Thereafter at each time
So the data are permanently generated as independently and identically distributed (IID) draws from either
We could say that objectively, meaning after nature has chosen either
We now drop into this setting a partially informed decision maker who
knows both
does not know whether at
Thus, although our decision maker knows
The decision maker describes his ignorance with a subjective probability
Thus, we assume that the decision maker
knows both
doesn’t know which of these two distributions that nature has drawn
expresses his ignorance by acting as if or thinking that nature chose distribution
at date
To proceed, we want to know the decision maker’s belief about the joint distribution of the partial history.
We’ll discuss that next and in the process describe the concept of exchangeability.
24.4. Relationship Between IID and Exchangeable#
Conditional on nature selecting
Conditional on nature selecting
Thus, conditional on nature having selected
Furthermore, conditional on nature having
selected
But what about the unconditional distribution of a partial history?
The unconditional distribution of
Under the unconditional distribution
To verify this claim, it is sufficient to notice, for example, that
Thus, the conditional distribution
This means that random variable
So there is something to learn from the past about the future.
24.5. Exchangeability#
While the sequence
and so on.
More generally, a sequence of random variables is said to be exchangeable if the joint probability distribution for a sequence does not change when the positions in the sequence in which finitely many of random variables appear are altered.
Equation (24.1) represents our instance of an exchangeable joint density over a sequence of random variables as a mixture of two IID joint densities over a sequence of random variables.
A Bayesian statistician interprets the mixing parameter
Note
DeFinetti [de Finetti, 1937] established a related representation of an exchangeable process created by mixing
sequences of IID Bernoulli random variables with parameter
24.6. Bayes’ Law#
We noted above that in our example model there is something to learn about about the future from past data drawn from our particular instance of a process that is exchangeable but not IID.
But how can we learn?
And about what?
The answer to the about what question is
The answer to the how question is to use Bayes’ Law.
Another way to say use Bayes’ Law is to say from a (subjective) joint distribution, compute an appropriate conditional distribution.
Let’s dive into Bayes’ Law in this context.
Let
where we regard
Suppose that at
We let
where we adopt the convention
The distribution of
Bayes’ rule for updating
Equation (24.2) follows from Bayes’ rule, which tells us that
where
24.7. More Details about Bayesian Updating#
Let’s stare at and rearrange Bayes’ Law as represented in equation (24.2) with the aim of understanding
how the posterior probability
It is convenient for us to rewrite the updating rule (24.2) as
This implies that
Notice how the likelihood ratio and the prior interact to determine whether an observation
When the likelihood ratio
Representation (24.3) is the foundation of some graphs that we’ll use to display the dynamics of
We’ll plot
To create the Python infrastructure to do our work for us, we construct a wrapper function that displays informative graphs
given parameters of
@vectorize
def p(x, a, b):
"The general beta distribution function."
r = gamma(a + b) / (gamma(a) * gamma(b))
return r * x ** (a-1) * (1 - x) ** (b-1)
def learning_example(F_a=1, F_b=1, G_a=3, G_b=1.2):
"""
A wrapper function that displays the updating rule of belief π,
given the parameters which specify F and G distributions.
"""
f = jit(lambda x: p(x, F_a, F_b))
g = jit(lambda x: p(x, G_a, G_b))
# l(w) = f(w) / g(w)
l = lambda w: f(w) / g(w)
# objective function for solving l(w) = 1
obj = lambda w: l(w) - 1
x_grid = np.linspace(0, 1, 100)
π_grid = np.linspace(1e-3, 1-1e-3, 100)
w_max = 1
w_grid = np.linspace(1e-12, w_max-1e-12, 100)
# the mode of beta distribution
# use this to divide w into two intervals for root finding
G_mode = (G_a - 1) / (G_a + G_b - 2)
roots = np.empty(2)
roots[0] = op.root_scalar(obj, bracket=[1e-10, G_mode]).root
roots[1] = op.root_scalar(obj, bracket=[G_mode, 1-1e-10]).root
fig, (ax1, ax2, ax3) = plt.subplots(1, 3, figsize=(18, 5))
ax1.plot(l(w_grid), w_grid, label='$l$', lw=2)
ax1.vlines(1., 0., 1., linestyle="--")
ax1.hlines(roots, 0., 2., linestyle="--")
ax1.set_xlim([0., 2.])
ax1.legend(loc=4)
ax1.set(xlabel='$l(w)=f(w)/g(w)$', ylabel='$w$')
ax2.plot(f(x_grid), x_grid, label='$f$', lw=2)
ax2.plot(g(x_grid), x_grid, label='$g$', lw=2)
ax2.vlines(1., 0., 1., linestyle="--")
ax2.hlines(roots, 0., 2., linestyle="--")
ax2.legend(loc=4)
ax2.set(xlabel='$f(w), g(w)$', ylabel='$w$')
area1 = quad(f, 0, roots[0])[0]
area2 = quad(g, roots[0], roots[1])[0]
area3 = quad(f, roots[1], 1)[0]
ax2.text((f(0) + f(roots[0])) / 4, roots[0] / 2, f"{area1: .3g}")
ax2.fill_between([0, 1], 0, roots[0], color='blue', alpha=0.15)
ax2.text(np.mean(g(roots)) / 2, np.mean(roots), f"{area2: .3g}")
w_roots = np.linspace(roots[0], roots[1], 20)
ax2.fill_betweenx(w_roots, 0, g(w_roots), color='orange', alpha=0.15)
ax2.text((f(roots[1]) + f(1)) / 4, (roots[1] + 1) / 2, f"{area3: .3g}")
ax2.fill_between([0, 1], roots[1], 1, color='blue', alpha=0.15)
W = np.arange(0.01, 0.99, 0.08)
Π = np.arange(0.01, 0.99, 0.08)
ΔW = np.zeros((len(W), len(Π)))
ΔΠ = np.empty((len(W), len(Π)))
for i, w in enumerate(W):
for j, π in enumerate(Π):
lw = l(w)
ΔΠ[i, j] = π * (lw / (π * lw + 1 - π) - 1)
q = ax3.quiver(Π, W, ΔΠ, ΔW, scale=2, color='r', alpha=0.8)
ax3.fill_between(π_grid, 0, roots[0], color='blue', alpha=0.15)
ax3.fill_between(π_grid, roots[0], roots[1], color='green', alpha=0.15)
ax3.fill_between(π_grid, roots[1], w_max, color='blue', alpha=0.15)
ax3.hlines(roots, 0., 1., linestyle="--")
ax3.set(xlabel='$\pi$', ylabel='$w$')
ax3.grid()
plt.show()
<>:78: SyntaxWarning: invalid escape sequence '\p'
<>:78: SyntaxWarning: invalid escape sequence '\p'
/tmp/ipykernel_6056/3701171347.py:78: SyntaxWarning: invalid escape sequence '\p'
ax3.set(xlabel='$\pi$', ylabel='$w$')
Now we’ll create a group of graphs that illustrate dynamics induced by Bayes’ Law.
We’ll begin with Python function default values of various objects, then change them in a subsequent example.
learning_example()
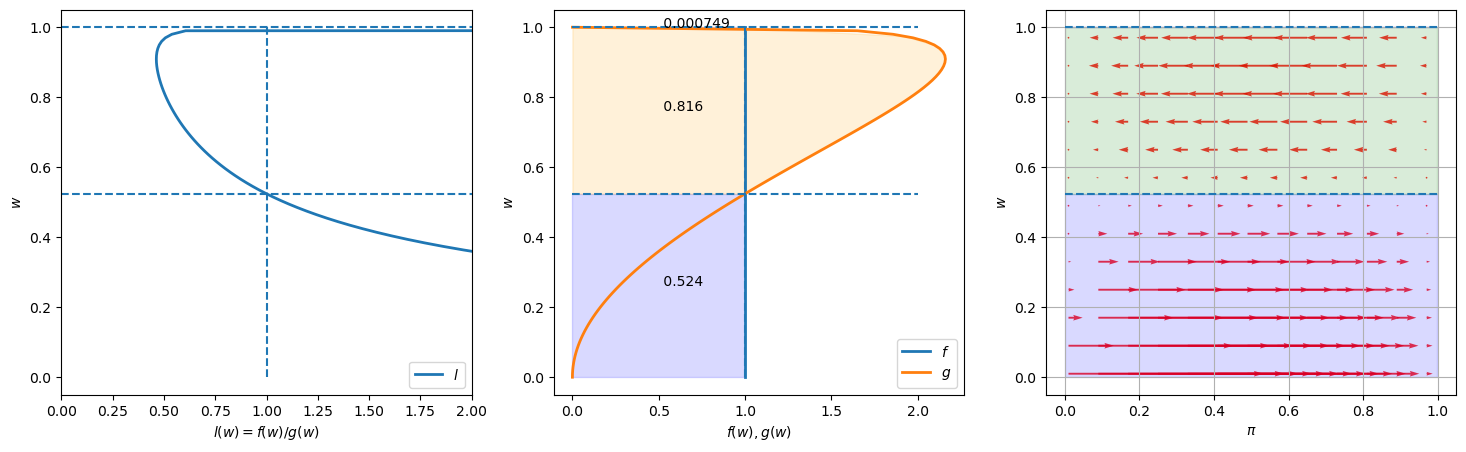
Please look at the three graphs above created for an instance in which
The graph on the left plots the likelihood ratio
The middle graph plots both
The graph on the right plots arrows to the right that show when Bayes’ Law makes
Lengths of the arrows show magnitudes of the force from Bayes’ Law impelling
These lengths depend on both the prior probability
The fractions in the colored areas of the middle graphs are probabilities under
For example,
in the above example, under true distribution
But this
would occur with probability
The fraction
Next we use our code to create graphs for another instance of our model.
We keep
learning_example(G_a=2, G_b=1.6)
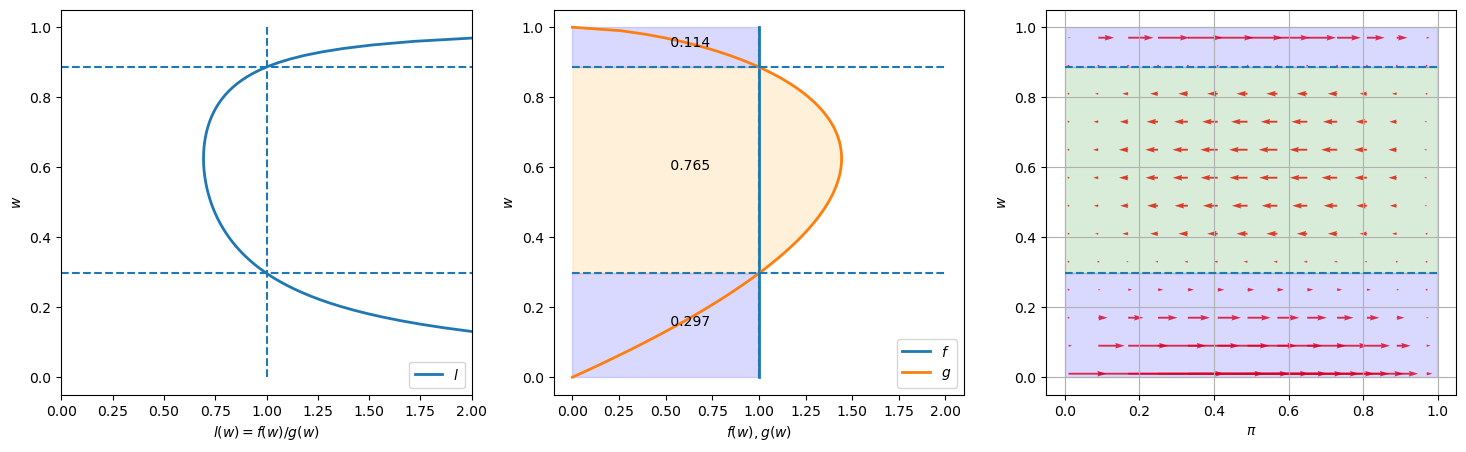
Notice how the likelihood ratio, the middle graph, and the arrows compare with the previous instance of our example.
24.8. Appendix#
24.8.1. Sample Paths of
Now we’ll have some fun by plotting multiple realizations of sample paths of
that nature permanently draws from
that nature permanently draws from
Outcomes depend on a peculiar property of likelihood ratio processes discussed in this lecture.
To proceed, we create some Python code.
def function_factory(F_a=1, F_b=1, G_a=3, G_b=1.2):
# define f and g
f = jit(lambda x: p(x, F_a, F_b))
g = jit(lambda x: p(x, G_a, G_b))
@jit
def update(a, b, π):
"Update π by drawing from beta distribution with parameters a and b"
# Draw
w = np.random.beta(a, b)
# Update belief
π = 1 / (1 + ((1 - π) * g(w)) / (π * f(w)))
return π
@jit
def simulate_path(a, b, T=50):
"Simulates a path of beliefs π with length T"
π = np.empty(T+1)
# initial condition
π[0] = 0.5
for t in range(1, T+1):
π[t] = update(a, b, π[t-1])
return π
def simulate(a=1, b=1, T=50, N=200, display=True):
"Simulates N paths of beliefs π with length T"
π_paths = np.empty((N, T+1))
if display:
fig = plt.figure()
for i in range(N):
π_paths[i] = simulate_path(a=a, b=b, T=T)
if display:
plt.plot(range(T+1), π_paths[i], color='b', lw=0.8, alpha=0.5)
if display:
plt.show()
return π_paths
return simulate
simulate = function_factory()
We begin by generating
T = 50
# when nature selects F
π_paths_F = simulate(a=1, b=1, T=T, N=1000)
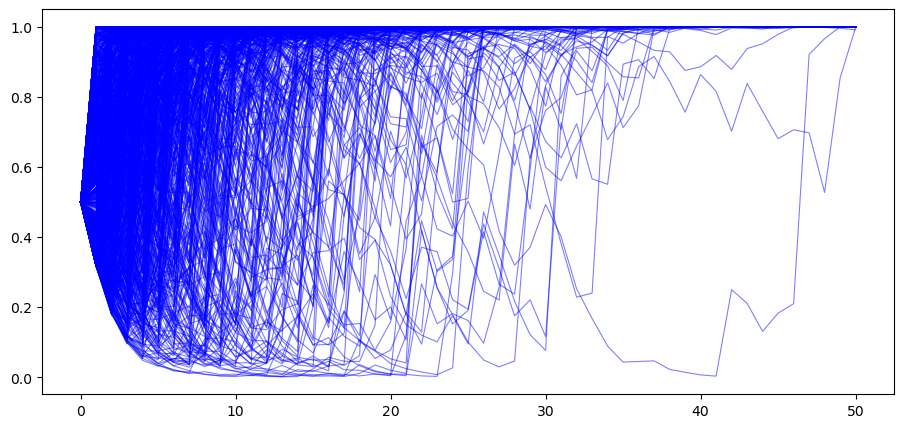
In the above example, for most paths
So Bayes’ Law evidently eventually discovers the truth for most of our paths.
Next, we generate paths with
# when nature selects G
π_paths_G = simulate(a=3, b=1.2, T=T, N=1000)
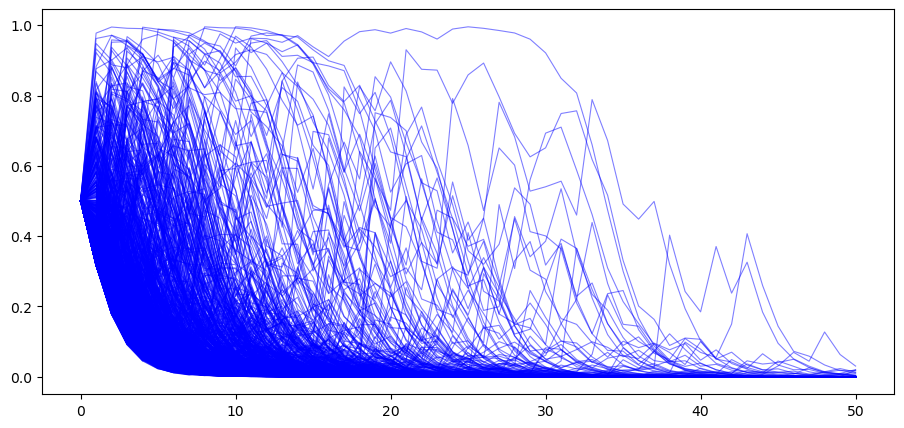
In the above graph we observe that now most paths
24.8.2. Rates of convergence#
We study rates of convergence of
We do this by averaging across simulated paths of
Using
plt.plot(range(T+1), 1 - np.mean(π_paths_F, 0), label='F generates')
plt.plot(range(T+1), np.mean(π_paths_G, 0), label='G generates')
plt.legend()
plt.title("convergence");
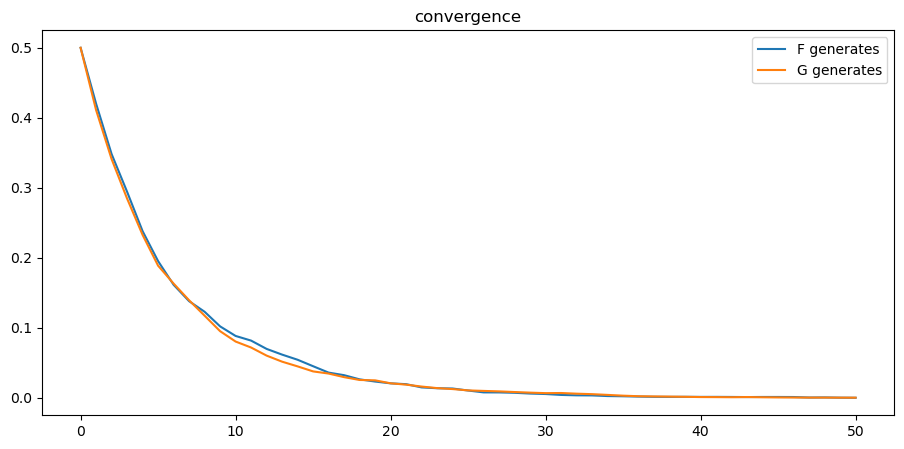
From the above graph, rates of convergence appear not to depend on whether
24.8.3. Graph of Ensemble Dynamics of
More insights about the dynamics of
where
The following code approximates the integral above:
def expected_ratio(F_a=1, F_b=1, G_a=3, G_b=1.2):
# define f and g
f = jit(lambda x: p(x, F_a, F_b))
g = jit(lambda x: p(x, G_a, G_b))
l = lambda w: f(w) / g(w)
integrand_f = lambda w, π: f(w) * l(w) / (π * l(w) + 1 - π)
integrand_g = lambda w, π: g(w) * l(w) / (π * l(w) + 1 - π)
π_grid = np.linspace(0.02, 0.98, 100)
expected_rario = np.empty(len(π_grid))
for q, inte in zip(["f", "g"], [integrand_f, integrand_g]):
for i, π in enumerate(π_grid):
expected_rario[i]= quad(inte, 0, 1, args=(π,))[0]
plt.plot(π_grid, expected_rario, label=f"{q} generates")
plt.hlines(1, 0, 1, linestyle="--")
plt.xlabel("$π_t$")
plt.ylabel("$E[\pi_{t+1}/\pi_t]$")
plt.legend()
plt.show()
<>:21: SyntaxWarning: invalid escape sequence '\p'
<>:21: SyntaxWarning: invalid escape sequence '\p'
/tmp/ipykernel_6056/2337625297.py:21: SyntaxWarning: invalid escape sequence '\p'
plt.ylabel("$E[\pi_{t+1}/\pi_t]$")
First, consider the case where
expected_ratio()
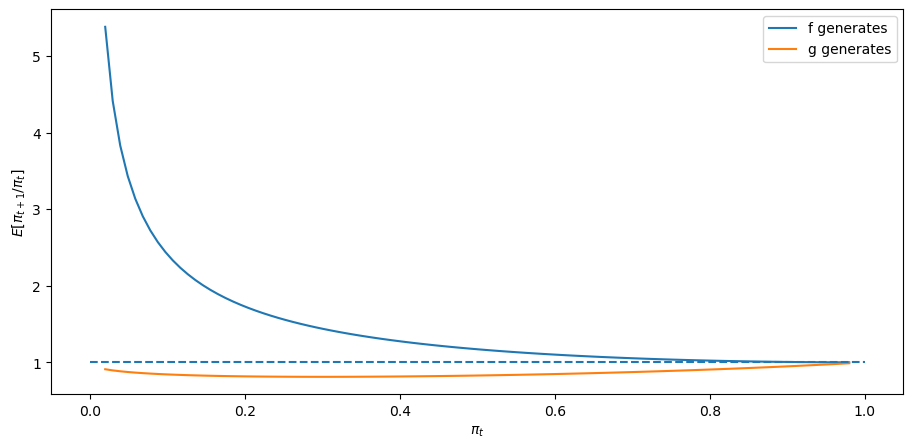
The above graphs shows that when
Next, we’ll look at a degenerate case in whcih
In a sense, here there is nothing to learn.
expected_ratio(F_a=3, F_b=1.2)
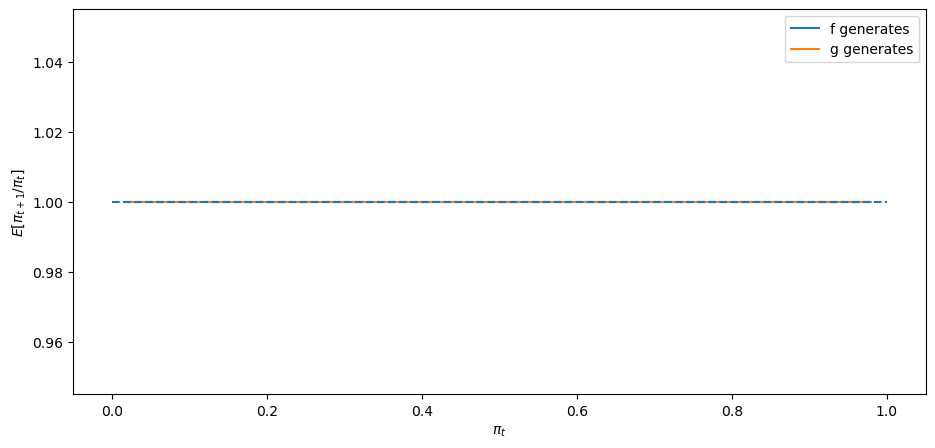
The above graph says that
Finally, let’s look at a case in which
expected_ratio(F_a=2, F_b=1, G_a=3, G_b=1.2)
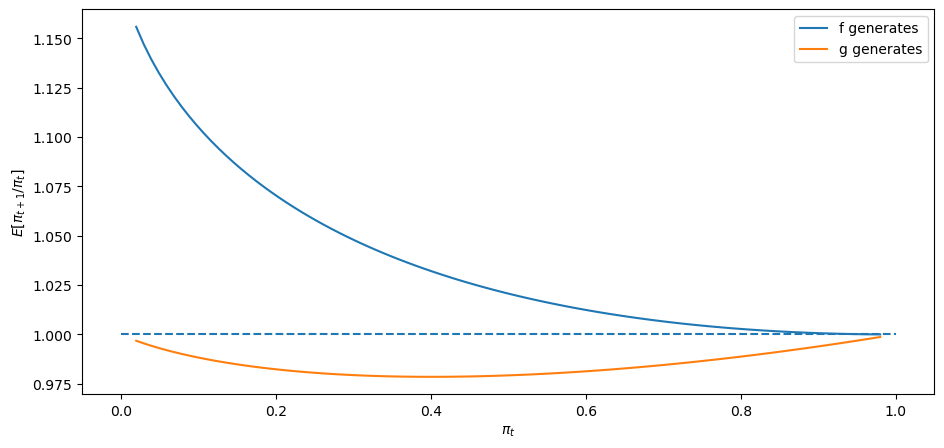
24.9. Sequels#
We’ll apply and dig deeper into some of the ideas presented in this lecture:
this lecture describes likelihood ratio processes and their role in frequentist and Bayesian statistical theories
this lecture studies whether a World War II US Navy Captain’s hunch that a (frequentist) decision rule that the Navy had told him to use was inferior to a sequential rule that Abraham Wald had not yet designed.