26. Incorrect Models#
GPU
This lecture was built using a machine with the latest CUDA and CUDANN frameworks installed with access to a GPU.
To run this lecture on Google Colab, click on the “play” icon top right, select Colab, and set the runtime environment to include a GPU.
To run this lecture on your own machine, you need to install the software listed following this notice.
!pip install numpyro jax
Show code cell output
Requirement already satisfied: numpyro in /home/runner/miniconda3/envs/quantecon/lib/python3.12/site-packages (0.18.0)
Requirement already satisfied: jax in /home/runner/miniconda3/envs/quantecon/lib/python3.12/site-packages (0.6.1)
Requirement already satisfied: jaxlib>=0.4.25 in /home/runner/miniconda3/envs/quantecon/lib/python3.12/site-packages (from numpyro) (0.6.1)
Requirement already satisfied: multipledispatch in /home/runner/miniconda3/envs/quantecon/lib/python3.12/site-packages (from numpyro) (0.6.0)
Requirement already satisfied: numpy in /home/runner/miniconda3/envs/quantecon/lib/python3.12/site-packages (from numpyro) (1.26.4)
Requirement already satisfied: tqdm in /home/runner/miniconda3/envs/quantecon/lib/python3.12/site-packages (from numpyro) (4.66.5)
Requirement already satisfied: ml_dtypes>=0.5.0 in /home/runner/miniconda3/envs/quantecon/lib/python3.12/site-packages (from jax) (0.5.1)
Requirement already satisfied: opt_einsum in /home/runner/miniconda3/envs/quantecon/lib/python3.12/site-packages (from jax) (3.4.0)
Requirement already satisfied: scipy>=1.11.1 in /home/runner/miniconda3/envs/quantecon/lib/python3.12/site-packages (from jax) (1.13.1)
Requirement already satisfied: six in /home/runner/miniconda3/envs/quantecon/lib/python3.12/site-packages (from multipledispatch->numpyro) (1.16.0)
26.1. Overview#
This is a sequel to this quantecon lecture.
We discuss two ways to create compound lottery and their consequences.
A compound lottery can be said to create a mixture distribution.
Our two ways of constructing a compound lottery will differ in their timing.
in one, mixing between two possible probability distributions will occur once and all at the beginning of time
in the other, mixing between the same two possible possible probability distributions will occur each period
The statistical setting is close but not identical to the problem studied in that quantecon lecture.
In that lecture, there were two i.i.d. processes that could possibly govern successive draws of a non-negative random variable
Nature decided once and for all whether to make a sequence of IID draws from either
That lecture studied an agent who knew both
The agent represented that ignorance by assuming that nature had chosen
That assumption allowed the agent to construct a subjective joint probability distribution over the
random sequence
We studied how the agent would then use the laws of conditional probability and an observed history
However, in the setting of this lecture, that rule imputes to the agent an incorrect model.
The reason is that now the wage sequence is actually described by a different statistical model.
Thus, we change the quantecon lecture specification in the following way.
Now, each period
Thus, naturally perpetually draws from the mixture distribution with c.d.f.
We’ll study two agents who try to learn about the wage process, but who use different statistical models.
Both types of agent know
Our first type of agent erroneously thinks that at time
Our second type of agent knows, correctly, that nature mixes
Our first type of agent applies the learning algorithm described in this quantecon lecture.
In the context of the statistical model that prevailed in that lecture, that was a good learning algorithm and it enabled the Bayesian learner
eventually to learn the distribution that nature had drawn at time
This is because the agent’s statistical model was correct in the sense of being aligned with the data generating process.
But in the present context, our type 1 decision maker’s model is incorrect because the model
Nevertheless, we’ll see that our first type of agent muddles through and eventually learns something interesting and useful, even though it is not true.
Instead, it turn out that our type 1 agent who is armed with a wrong statistical model ends up learning whichever probability distribution,
We’ll tell the sense in which it is closest.
Our second type of agent understands that nature mixes between
But the agent doesn’t know
The agent sets out to learn
His model is correct in the sense that
it includes the actual data generating process
In this lecture, we’ll learn about
how nature can mix between two distributions
The Kullback-Leibler statistical divergence https://en.wikipedia.org/wiki/Kullback–Leibler_divergence that governs statistical learning under an incorrect statistical model
A useful Python function
numpy.searchsortedthat, in conjunction with a uniform random number generator, can be used to sample from an arbitrary distribution
As usual, we’ll start by importing some Python tools.
import matplotlib.pyplot as plt
plt.rcParams["figure.figsize"] = (11, 5) #set default figure size
import numpy as np
from numba import vectorize, jit
from math import gamma
import pandas as pd
import scipy.stats as sp
from scipy.integrate import quad
import seaborn as sns
colors = sns.color_palette()
import numpyro
import numpyro.distributions as dist
from numpyro.infer import MCMC, NUTS
import jax.numpy as jnp
from jax import random
np.random.seed(142857)
@jit
def set_seed():
np.random.seed(142857)
set_seed()
Let’s use Python to generate two beta distributions
# Parameters in the two beta distributions.
F_a, F_b = 1, 1
G_a, G_b = 3, 1.2
@vectorize
def p(x, a, b):
r = gamma(a + b) / (gamma(a) * gamma(b))
return r * x** (a-1) * (1 - x) ** (b-1)
# The two density functions.
f = jit(lambda x: p(x, F_a, F_b))
g = jit(lambda x: p(x, G_a, G_b))
@jit
def simulate(a, b, T=50, N=500):
'''
Generate N sets of T observations of the likelihood ratio,
return as N x T matrix.
'''
l_arr = np.empty((N, T))
for i in range(N):
for j in range(T):
w = np.random.beta(a, b)
l_arr[i, j] = f(w) / g(w)
return l_arr
We’ll also use the following Python code to prepare some informative simulations
l_arr_g = simulate(G_a, G_b, N=50000)
l_seq_g = np.cumprod(l_arr_g, axis=1)
l_arr_f = simulate(F_a, F_b, N=50000)
l_seq_f = np.cumprod(l_arr_f, axis=1)
26.2. Sampling from Compound Lottery
We implement two methods to draw samples from
our mixture model
We’ll generate samples using each of them and verify that they match well.
Here is pseudo code for a direct “method 1” for drawing from our compound lottery:
Step one:
use the numpy.random.choice function to flip an unfair coin that selects distribution
Step two:
draw from either
Step three:
put the first two steps in a big loop and do them for each realization of
Our second method uses a uniform distribution and the following fact that we also described and used in the quantecon lecture https://python.quantecon.org/prob_matrix.html:
If a random variable
In other words, if
We’ll use this fact
in conjunction with the numpy.searchsorted command to sample from
See https://numpy.org/doc/stable/reference/generated/numpy.searchsorted.html for the
searchsorted function.
See the Mr. P Solver video on Monte Carlo simulation to see other applications of this powerful trick.
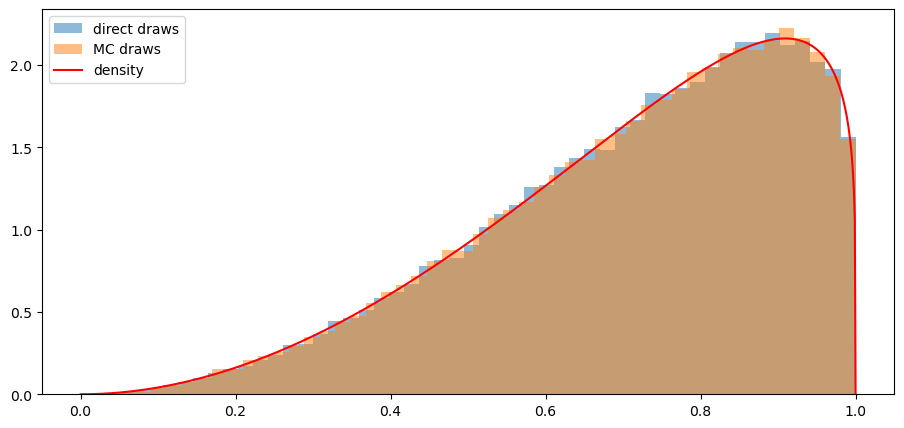
In the Python code below, we’ll use both of our methods and confirm that each of them does a good job of sampling from our target mixture distribution.
@jit
def draw_lottery(p, N):
"Draw from the compound lottery directly."
draws = []
for i in range(0, N):
if np.random.rand()<=p:
draws.append(np.random.beta(F_a, F_b))
else:
draws.append(np.random.beta(G_a, G_b))
return np.array(draws)
def draw_lottery_MC(p, N):
"Draw from the compound lottery using the Monte Carlo trick."
xs = np.linspace(1e-8,1-(1e-8),10000)
CDF = p*sp.beta.cdf(xs, F_a, F_b) + (1-p)*sp.beta.cdf(xs, G_a, G_b)
Us = np.random.rand(N)
draws = xs[np.searchsorted(CDF[:-1], Us)]
return draws
# verify
N = 100000
α = 0.0
sample1 = draw_lottery(α, N)
sample2 = draw_lottery_MC(α, N)
# plot draws and density function
plt.hist(sample1, 50, density=True, alpha=0.5, label='direct draws')
plt.hist(sample2, 50, density=True, alpha=0.5, label='MC draws')
xs = np.linspace(0,1,1000)
plt.plot(xs, α*f(xs)+(1-α)*g(xs), color='red', label='density')
plt.legend()
plt.show()
# %%timeit # compare speed
# sample1 = draw_lottery(α, N=int(1e6))
# %%timeit
# sample2 = draw_lottery_MC(α, N=int(1e6))
Note: With numba acceleration the first method is actually only slightly slower than the second when we generated 1,000,000 samples.
26.3. Type 1 Agent#
We’ll now study what our type 1 agent learns
Remember that our type 1 agent uses the wrong statistical model, thinking that nature mixed between
The type 1 agent thus uses the learning algorithm studied in this quantecon lecture.
We’ll briefly review that learning algorithm now.
Let
The likelihood ratio process plays a principal role in the formula that governs the evolution
of the posterior probability
Bayes’ law implies that
with
Below we define a Python function that updates belief
@jit
def update(π, l):
"Update π using likelihood l"
# Update belief
π = π * l / (π * l + 1 - π)
return π
Formula (26.1) can be generalized by iterating on it and thereby deriving an
expression for the time
To begin, notice that the updating rule
implies
Therefore
Since
After rearranging the preceding equation, we can express
Formula (26.2) generalizes formula (26.1).
Formula (26.2) can be regarded as a one step revision of prior probability
26.4. What a type 1 Agent Learns when Mixture
We now study what happens when the mixture distribution
A submartingale or supermartingale continues to describe
It raises its ugly head and causes
This is true even though in truth nature always mixes between
After verifying that claim about possible limit points of
Let’s set a value of
def simulate_mixed(α, T=50, N=500):
"""
Generate N sets of T observations of the likelihood ratio,
return as N x T matrix, when the true density is mixed h;α
"""
w_s = draw_lottery(α, N*T).reshape(N, T)
l_arr = f(w_s) / g(w_s)
return l_arr
def plot_π_seq(α, π1=0.2, π2=0.8, T=200):
"""
Compute and plot π_seq and the log likelihood ratio process
when the mixed distribution governs the data.
"""
l_arr_mixed = simulate_mixed(α, T=T, N=50)
l_seq_mixed = np.cumprod(l_arr_mixed, axis=1)
T = l_arr_mixed.shape[1]
π_seq_mixed = np.empty((2, T+1))
π_seq_mixed[:, 0] = π1, π2
for t in range(T):
for i in range(2):
π_seq_mixed[i, t+1] = update(π_seq_mixed[i, t], l_arr_mixed[0, t])
# plot
fig, ax1 = plt.subplots()
for i in range(2):
ax1.plot(range(T+1), π_seq_mixed[i, :], label=rf"$\pi_0$={π_seq_mixed[i, 0]}")
ax1.plot(np.nan, np.nan, '--', color='b', label='Log likelihood ratio process')
ax1.set_ylabel("$\pi_t$")
ax1.set_xlabel("t")
ax1.legend()
ax1.set_title("when $\\alpha G + (1-\\alpha)$ F governs data")
ax2 = ax1.twinx()
ax2.plot(range(1, T+1), np.log(l_seq_mixed[0, :]), '--', color='b')
ax2.set_ylabel("$log(L(w^{t}))$")
plt.show()
<>:35: SyntaxWarning: invalid escape sequence '\p'
<>:35: SyntaxWarning: invalid escape sequence '\p'
/tmp/ipykernel_7565/2013898715.py:35: SyntaxWarning: invalid escape sequence '\p'
ax1.set_ylabel("$\pi_t$")
plot_π_seq(α = 0.6)
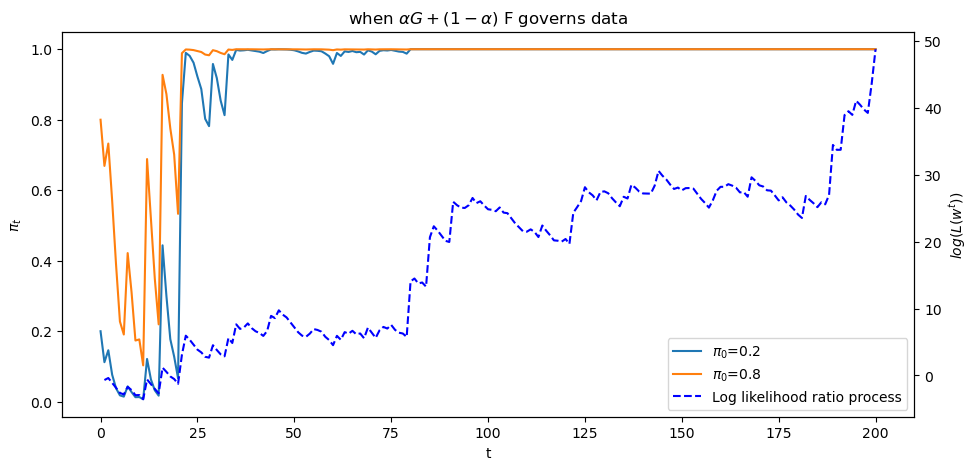
The above graph shows a sample path of the log likelihood ratio process as the blue dotted line, together with
sample paths of
Let’s see what happens when we change
plot_π_seq(α = 0.2)
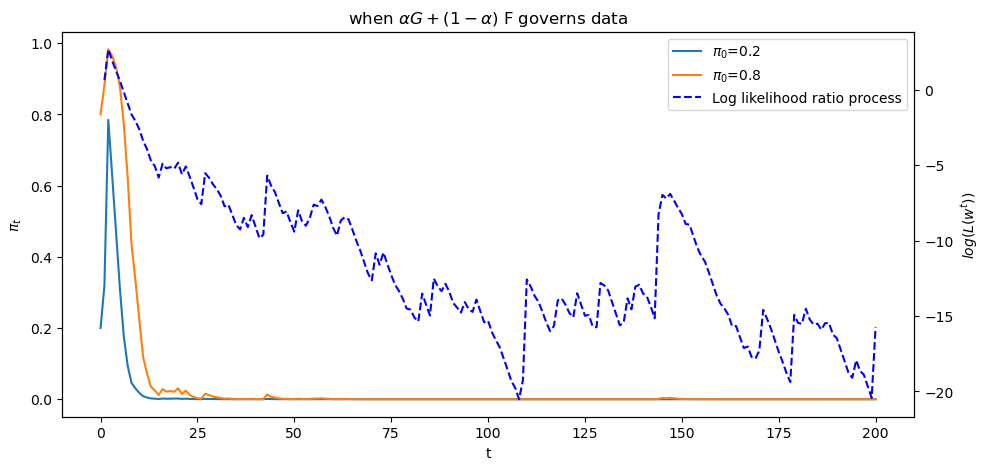
Evidently,
26.5. Kullback-Leibler Divergence Governs Limit of
To understand what determines whether the limit point of
we shall compute the following two Kullback-Leibler divergences
and
We shall plot both of these functions against
The limit of
The only possible limits are
As
@vectorize
def KL_g(α):
"Compute the KL divergence between g and h."
err = 1e-8 # to avoid 0 at end points
ws = np.linspace(err, 1-err, 10000)
gs, fs = g(ws), f(ws)
hs = α*fs + (1-α)*gs
return np.sum(np.log(gs/hs)*hs)/10000
@vectorize
def KL_f(α):
"Compute the KL divergence between f and h."
err = 1e-8 # to avoid 0 at end points
ws = np.linspace(err, 1-err, 10000)
gs, fs = g(ws), f(ws)
hs = α*fs + (1-α)*gs
return np.sum(np.log(fs/hs)*hs)/10000
# compute KL using quad in Scipy
def KL_g_quad(α):
"Compute the KL divergence between g and h using scipy.integrate."
h = lambda x: α*f(x) + (1-α)*g(x)
return quad(lambda x: np.log(g(x)/h(x))*h(x), 0, 1)[0]
def KL_f_quad(α):
"Compute the KL divergence between f and h using scipy.integrate."
h = lambda x: α*f(x) + (1-α)*g(x)
return quad(lambda x: np.log(f(x)/h(x))*h(x), 0, 1)[0]
# vectorize
KL_g_quad_v = np.vectorize(KL_g_quad)
KL_f_quad_v = np.vectorize(KL_f_quad)
# Let us find the limit point
def π_lim(α, T=5000, π_0=0.4):
"Find limit of π sequence."
π_seq = np.zeros(T+1)
π_seq[0] = π_0
l_arr = simulate_mixed(α, T, N=1)[0]
for t in range(T):
π_seq[t+1] = update(π_seq[t], l_arr[t])
return π_seq[-1]
π_lim_v = np.vectorize(π_lim)
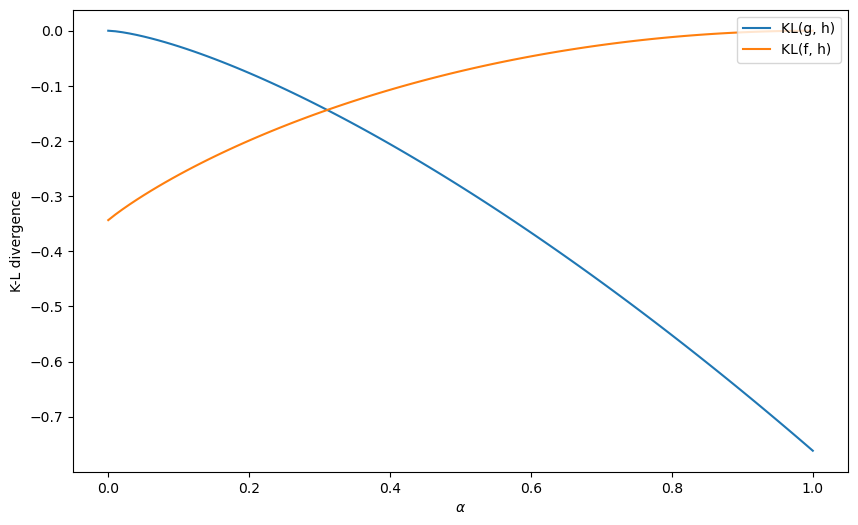
Let us first plot the KL divergences
α_arr = np.linspace(0, 1, 100)
KL_g_arr = KL_g(α_arr)
KL_f_arr = KL_f(α_arr)
fig, ax = plt.subplots(1, figsize=[10, 6])
ax.plot(α_arr, KL_g_arr, label='KL(g, h)')
ax.plot(α_arr, KL_f_arr, label='KL(f, h)')
ax.set_ylabel('K-L divergence')
ax.set_xlabel(r'$\alpha$')
ax.legend(loc='upper right')
plt.show()
# # using Scipy to compute KL divergence
# α_arr = np.linspace(0, 1, 100)
# KL_g_arr = KL_g_quad_v(α_arr)
# KL_f_arr = KL_f_quad_v(α_arr)
# fig, ax = plt.subplots(1, figsize=[10, 6])
# ax.plot(α_arr, KL_g_arr, label='KL(g, h)')
# ax.plot(α_arr, KL_f_arr, label='KL(f, h)')
# ax.set_ylabel('K-L divergence')
# ax.legend(loc='upper right')
# plt.show()
Let’s compute an
# where KL_f = KL_g
α_arr[np.argmin(np.abs(KL_g_arr-KL_f_arr))]
0.31313131313131315
We can compute and plot the convergence point
The blue circles show the limiting values of
Thus, the graph below confirms how a minimum KL divergence governs what our type 1 agent eventually learns.
α_arr_x = α_arr[(α_arr<0.28)|(α_arr>0.38)]
π_lim_arr = π_lim_v(α_arr_x)
# plot
fig, ax = plt.subplots(1, figsize=[10, 6])
ax.plot(α_arr, KL_g_arr, label='KL(g, h)')
ax.plot(α_arr, KL_f_arr, label='KL(f, h)')
ax.set_ylabel('K-L divergence')
ax.set_xlabel(r'$\alpha$')
# plot KL
ax2 = ax.twinx()
# plot limit point
ax2.scatter(α_arr_x, π_lim_arr, facecolors='none', edgecolors='tab:blue', label=r'$\pi$ lim')
ax2.set_ylabel('π lim')
ax.legend(loc=[0.85, 0.8])
ax2.legend(loc=[0.85, 0.73])
plt.show()
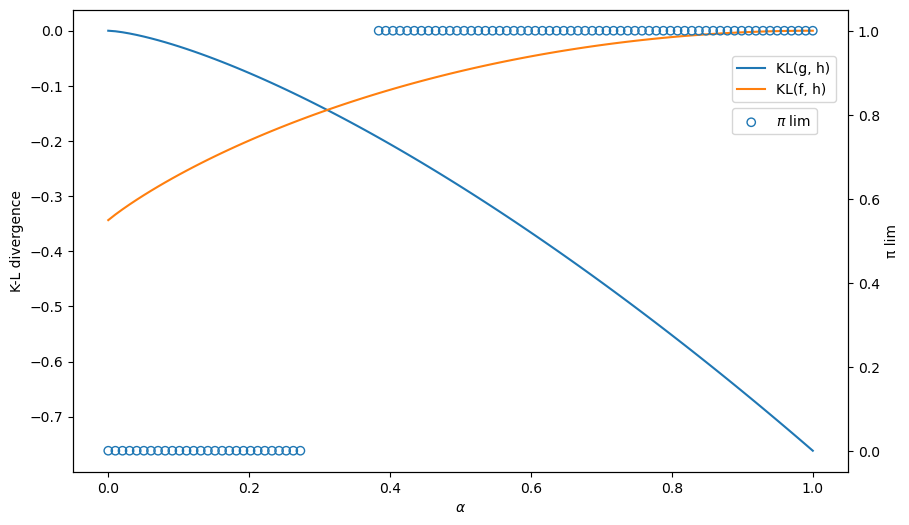
Evidently, our type 1 learner who applies Bayes’ law to his misspecified set of statistical models eventually learns an approximating model that is as close as possible to the true model, as measured by its Kullback-Leibler divergence.
26.6. Type 2 Agent#
We now describe how our type 2 agent formulates his learning problem and what he eventually learns.
Our type 2 agent understands the correct statistical model but acknowledges does not know
We apply Bayes law to deduce an algorithm for learning
but does not know
We’ll assume that the person starts out with a prior probabilty
We’ll fire up numpyro and apply it to the present situation.
Bayes’ law now takes the form
We’ll use numpyro to approximate this equation.
We’ll create graphs of the posterior
We anticipate that a posterior distribution will collapse around the true
Let us try a uniform prior first.
We use the Mixture class in Numpyro to construct the likelihood function.
α = 0.8
# simulate data with true α
data = draw_lottery(α, 1000)
sizes = [5, 20, 50, 200, 1000, 25000]
def model(w):
α = numpyro.sample('α', dist.Uniform(low=0.0, high=1.0))
y_samp = numpyro.sample('w',
dist.Mixture(dist.Categorical(jnp.array([α, 1-α])), [dist.Beta(F_a, F_b), dist.Beta(G_a, G_b)]), obs=w)
def MCMC_run(ws):
"Compute posterior using MCMC with observed ws"
kernal = NUTS(model)
mcmc = MCMC(kernal, num_samples=5000, num_warmup=1000, progress_bar=False)
mcmc.run(rng_key=random.PRNGKey(142857), w=jnp.array(ws))
sample = mcmc.get_samples()
return sample['α']
The following code generates the graph below that displays Bayesian posteriors for
fig, ax = plt.subplots(figsize=(10, 6))
for i in range(len(sizes)):
sample = MCMC_run(data[:sizes[i]])
sns.histplot(
data=sample, kde=True, stat='density', alpha=0.2, ax=ax,
color=colors[i], binwidth=0.02, linewidth=0.05, label=f't={sizes[i]}'
)
ax.set_title(r'$\pi_t(\alpha)$ as $t$ increases')
ax.legend()
ax.set_xlabel(r'$\alpha$')
plt.show()
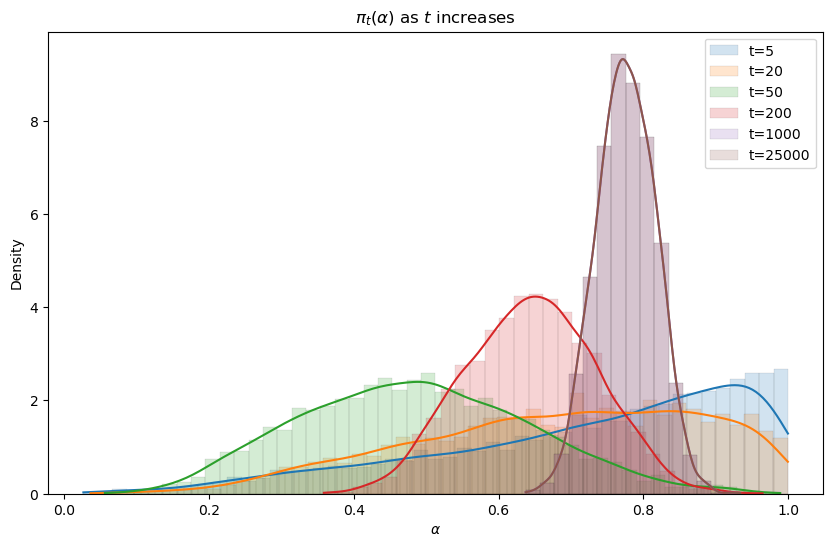
Evidently, the Bayesian posterior narrows in on the true value
26.7. Concluding Remarks#
Our type 1 person deploys an incorrect statistical model.
He believes
that either
That is wrong because nature is actually mixing each period with mixing probability
Our type 1 agent eventually believes that either
Our type 2 agent has a different statistical model, one that is correctly specified.
He knows the parametric form of the statistical model but not the mixing parameter
He knows that he does not know it.
But by using Bayes’ law in conjunction with his statistical model and a history of data, he eventually acquires a more and more accurate inference about
This little laboratory exhibits some important general principles that govern outcomes of Bayesian learning of misspecified models.
Thus, the following situation prevails quite generally in empirical work.
A scientist approaches the data with a manifold
The scientist with observations that he interprests as realizations
But the scientist’s model is misspecified, being only an approximation to an unknown model
If the scientist uses Bayes’ law or a related likelihood-based method to infer